Making Julia as Fast as C++
18 Apr 2019, Eduardo Alvarez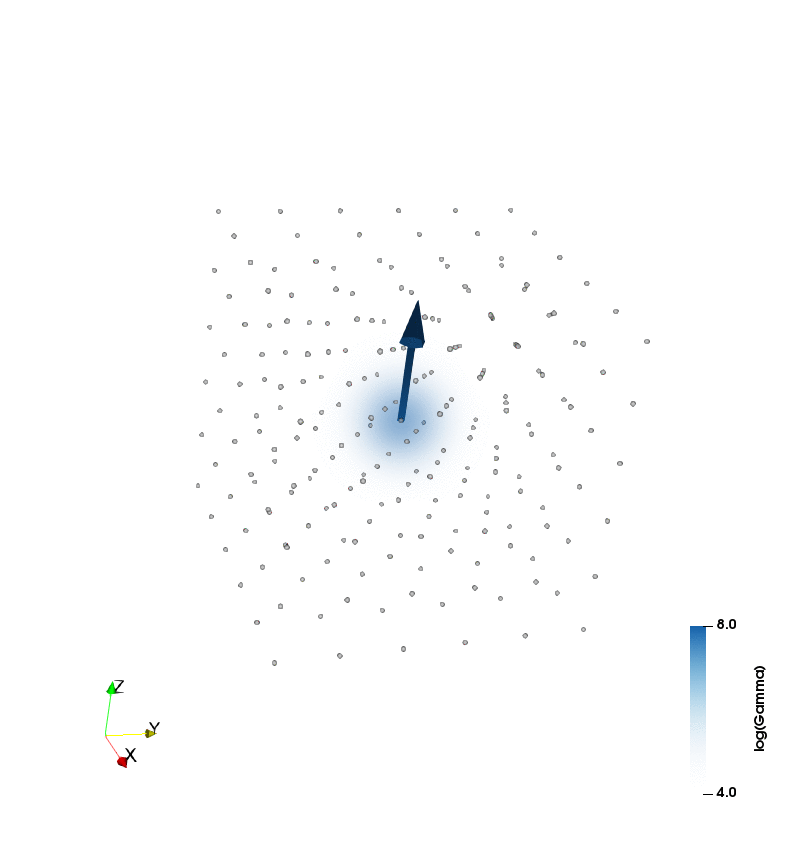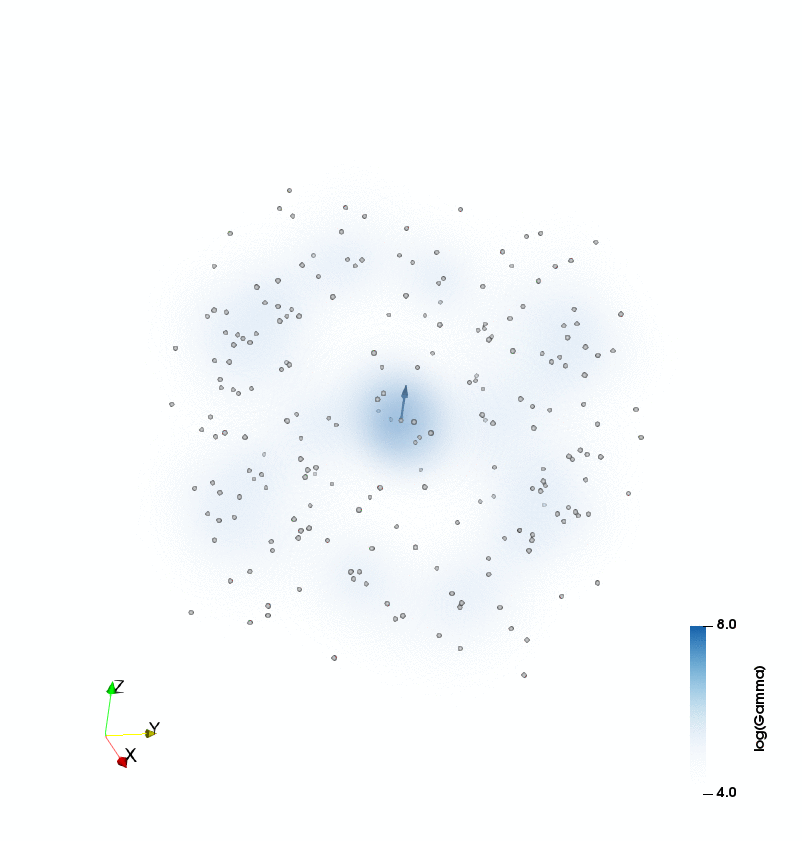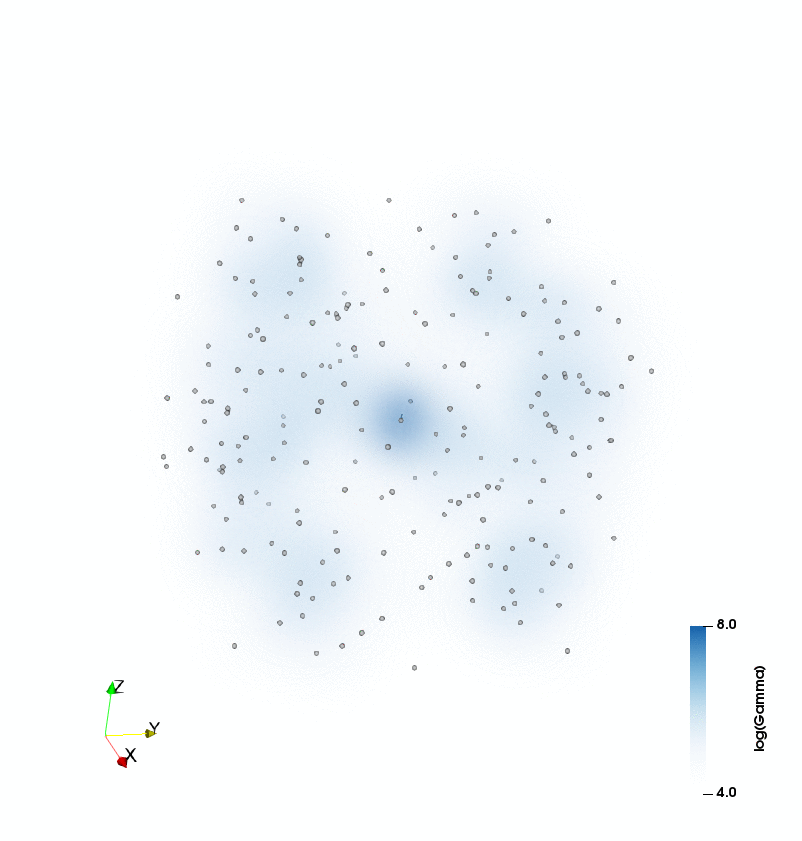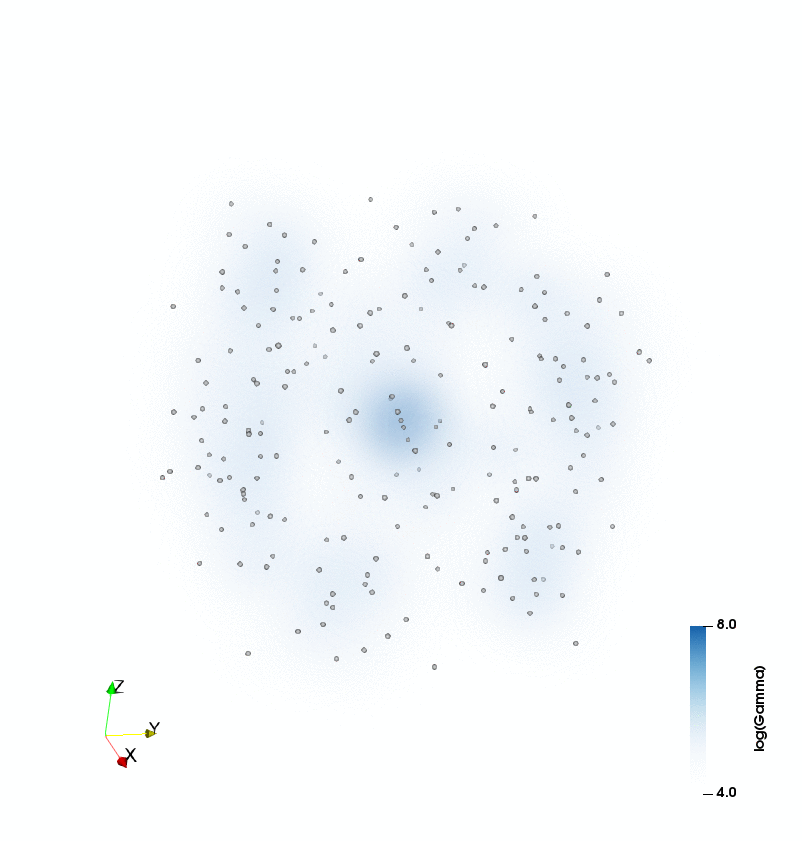
Introduction
The rumor says that Julia can achieve the same computing performance as any other compiled language like C++ and FORTRAN. After coding in Julia for the past two years I have definitely fell in love with its pythonic syntax, multiple dispatch, and MATLAB-like handiness in linear algebra, while being able to use compilation features like explicit type declaration for bug-preventive programming. In summary, Julia’s philosophy brings all the flexibility of an interpreted language, meanwhile its Just-In-Time (JIT) compilation makes it a defacto compiled language.
Julia’s high level syntax makes the language easygoing for programmers from any background, however, achieving high performance is sort of an art. In this post I summarize some of the things I’ve learned while crafting my Julia codes for high-performance computing. I will attempt to show the process of code optimization through a real-world computing application from aerodynamics: the vortex particle method\(^{[1,\,2]}\).
Problem Definition
In the vortex particle method we are interested in calculating the velocity \(\mathbf{u}\) and velocity Jacobian \(\mathbf{J}\) that a field of \(N\) vortex particles induces at an arbitrary position \(\mathbf{x}\). This is calculated as
\[{\bf u}\left( {\bf x} \right) = \sum\limits_p^N g_\sigma\left( {\bf x}-{\bf x}_p \right) {\bf K}\left( {\bf x}-{\bf x}_p \right) \times \boldsymbol\Gamma_p\] \[\frac{\partial {\bf u}}{\partial x_j}\left( {\bf x} \right) = \sum\limits_p^N \left[ \left( \frac{1}{\sigma } \frac{\Delta x_j}{\Vert \Delta \mathbf{x} \Vert} \frac{\partial g}{\partial r} \left( \frac{\Vert \Delta\mathbf{x} \Vert}{\sigma} \right) - 3 g_\sigma\left( \Delta{\bf x} \right) \frac{\Delta x_j}{\Vert \Delta\mathbf{x} \Vert^2} \right) {\bf K}\left( \Delta\mathbf{x} \right) \times \boldsymbol\Gamma_p - \frac{g_\sigma\left( \Delta{\bf x} \right) }{4\pi \Vert \Delta{\bf x} \Vert^3} \delta_{ij} \times \boldsymbol\Gamma_p \right] ,\]with \({\bf K}\) the singular Newtonian kernel \({\bf K}\left( {\bf x}\right)=-\frac{ {\bf x} }{4\pi \Vert{\bf x}\Vert^3}\), \(g_\sigma\) a regularizing function of smoothing radius \(\sigma\), and \(\mathbf{x}_p\) and \(\boldsymbol{\Gamma}_p\) the position and vectorial strength of the \(p\)-th particle, respectively. Furthermore, the governing equations of the method require evaluating the velocity \(\mathbf{u}\) and Jacobian \(\mathbf{J}\) that the collection of particles induces on itself, leading to the well-known \(N\)-body problem of computational complexity \(\mathcal{O}(N^2)\).
Choosing Winckelmans’ regularizing kernel\(^{[1]}\)
\[g(r) = r^3 \frac{r^2 + 5/2}{\left( r^2 + 1 \right)^{5/2}}\] \[\frac{\partial g}{\partial r} (r) = \frac{15}{2} \frac{r^2}{\left( r^2 + 1 \right)^{7/2}} ,\]the above equations are implemented in C++ as follows:
// Particle-to-particle interactions
void P2P(Particle * P, const int numParticles) {
real_t r, ros, aux, g_sgm, dgdr;
vec3 dX;
for (int i=0; i<numParticles; i++) {
for (int j=0; j<numParticles; j++) {
dX[0] = P[i].X[0] - P[j].X[0];
dX[1] = P[i].X[1] - P[j].X[1];
dX[2] = P[i].X[2] - P[j].X[2];
r = sqrt(dX[0]*dX[0] + dX[1]*dX[1] + dX[2]*dX[2]);
if (r!=0){
ros = r/P[j].sigma;
// Evaluate g_σ and ∂gσ∂r
aux = pow(ros*ros + 1.0, 2.5);
g_sgm = ros*ros*ros * (ros*ros + 2.5) / aux;
dgdr = 7.5 * ros*ros / ( aux * (ros*ros + 1.0) );
// u(x) = ∑g_σ(x−xp) K(x−xp) × Γp
aux = (- const4 / (r*r*r)) * g_sgm;
P[i].U[0] += ( dX[1]*P[j].Gamma[2] - dX[2]*P[j].Gamma[1] ) * aux;
P[i].U[1] += ( dX[2]*P[j].Gamma[0] - dX[0]*P[j].Gamma[2] ) * aux;
P[i].U[2] += ( dX[0]*P[j].Gamma[1] - dX[1]*P[j].Gamma[0] ) * aux;
// ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp]
aux = (- const4 / (r*r*r)) * (dgdr/(P[j].sigma*r) - 3.0*g_sgm/(r*r));
for (int k=0; k<3; k++){
P[i].J[3*k + 0] += ( dX[1]*P[j].Gamma[2] - dX[2]*P[j].Gamma[1] )*aux*dX[k];
P[i].J[3*k + 1] += ( dX[2]*P[j].Gamma[0] - dX[0]*P[j].Gamma[2] )*aux*dX[k];
P[i].J[3*k + 2] += ( dX[0]*P[j].Gamma[1] - dX[1]*P[j].Gamma[0] )*aux*dX[k];
}
// Adds the Kronecker delta term
aux = - const4 * g_sgm / (r*r*r);
// k=1
P[i].J[3*0 + 1] -= aux * P[j].Gamma[2];
P[i].J[3*0 + 2] += aux * P[j].Gamma[1];
// k=2
P[i].J[3*1 + 0] += aux * P[j].Gamma[2];
P[i].J[3*1 + 2] -= aux * P[j].Gamma[0];
// k=3
P[i].J[3*2 + 0] -= aux * P[j].Gamma[1];
P[i].J[3*2 + 1] += aux * P[j].Gamma[0];
}
}
}
}
The figure at the top of the page is a simulation of a 6x6x6 box of particles with vorticity initially concentrated at its center, diffusing as the simulation progresses due to viscous effects:
C++ Benchmark
Here I have coded a benchmark
of the C++ code shown above, evaluating P2P on a box of 6x6x6=216 particles,
and made it callable in this notebook through the
CxxWrap Julia package:
In [1]:
#=
This cell loads auxiliary benchmarking functions available
here: https://github.com/EdoAlvarezR/post-juliavscpp
=#
using LinearAlgebra
# Load C++ code wrapped as module CxxVortexTest
include("code/vortextest.jl")
# Load benchmarking tools
include("code/benchmarking.jl");In [2]:
ntests = 1000 # Tests to run
n = 6 # Particles per side
lambda = 1.5 # Core overlap
verbose = true
# Run C++ benchmark
cpptime = CxxVortexTest.benchmarkP2P_wrap(ntests, n, Float32(lambda), verbose)
# Store benchmark result
benchtime["C++"] = cpptime;Samples: 1000
min time: 3.99555 ms
ave time: 4.77778 ms
max time: 6.73414 ms
This was ran in my Dell Latitude 5580 laptop (Intel® Core™ i7-7820HQ CPU @ 2.90GHz × 8 ) in only one process, and we see that the C++ kernel, best-case scenario, is evaluated in ~4 ms. Let’s move on and code this up in Julia.
NOTE: The C++ code was compiled with the -O3 flag for code optimization.
Optimizing Julia
Julia Baseline: Pythonic Programming
Tempted by the Python-like syntax available in Julia, our first inclination is to make the code as general, simple, and easy to understand as possible. Here is the most general implementation where no types are specified:
In [3]:
"""
This is a particle struct made up of ambiguous (unspecified) types
"""
struct ParticleAmbiguous
# User inputs
X # Position
Gamma # Vectorial circulation
sigma # Smoothing radius
# Properties
U # Velocity at particle
J # Jacobian at particle J[i,j]=dUi/dxj
ParticleAmbiguous(X, Gamma, sigma; U=zeros(3), J=zeros(3,3)
) = new(X, Gamma, sigma, U, J )
end
# Empty initializer
Base.zero(::Type{<:ParticleAmbiguous}) = ParticleAmbiguous(zeros(3), zeros(3), 0.0)
# Winckelmans regularizing kernel
g_wnk(r) = r^3 * (r^2 + 2.5) / (r^2 + 1)^2.5
dgdr_wnk(r) = 7.5 * r^2 / (r^2 + 1)^3.5
const const4 = 1/(4*pi)
"""
Pythonic programming approach
"""
function P2P_pythonic(particles, g, dgdr)
for Pi in particles
for Pj in particles
dX = Pi.X - Pj.X
r = norm(dX)
if r != 0
# g_σ and ∂gσ∂r
gsgm = g(r / Pj.sigma)
dgsgmdr = dgdr(r / Pj.sigma)
# K × Γp
crss = cross(-const4 * (dX/r^3), Pj.Gamma)
# U = ∑g_σ(x-xp) * K(x-xp) × Γp
Pi.U[:] += gsgm * crss
# ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp ]
for j in 1:3
Pi.J[:, j] += ( dX[j] / (Pj.sigma*r) * (dgsgmdr*crss) -
gsgm * (3*dX[j]/r^2) * crss -
gsgm * (const4/r^3) * cross([i==j for i in 1:3], Pj.Gamma) )
end
end
end
end
endIn [4]:
# Create a 6x6x6 box of ambiguous particles
particles_amb = generate_particles(ParticleAmbiguous, n, lambda)
# Run benchmark
args = (particles_amb, g_wnk, dgdr_wnk)
compare(P2P_pythonic, "C++", args; reverse=false)
@benchmark P2P_pythonic(args...);C++ is 58.48 times faster than P2P_pythonic (3.996ms vs 233.679ms)
BenchmarkTools.Trial:
memory estimate: 217.57 MiB
allocs estimate: 4087152
--------------
minimum time: 233.679 ms (5.04% GC)
median time: 238.482 ms (4.99% GC)
mean time: 251.540 ms (9.28% GC)
maximum time: 295.517 ms (19.89% GC)
--------------
samples: 4
evals/sample: 1
Here we see that in our pythonic attempt we’ve got code that is pretty neat and concise, but ~58x slower than the C++ implementation.
Fix #1: Always work with concrete types
The problem with the pythonic approach is that variable types are never
declared, and without foreknowledge of the types to be handled, Julia can’t
optimize the function during compilation. In order to help us catch ambiguous
(abstract) types in our code, the Julia Base package provides the macro
@code_warntype, which prints the lowered and type-inferred AST used during
compilation highlighting any abstract types encountered.
The output is pretty lengthy, so here I have copied and pasted only a snippet:
@code_warntype P2P_pythonic(args...)
Body::Nothing
│╻╷╷ iterate34 1 ── %1 = (Base.arraylen)(particles)::Int64
││╻╷ iterate │ %2 = (Base.sle_int)(0, %1)::Bool
│││╻ < │ %3 = (Base.bitcast)(UInt64, %1)::UInt64
││││╻ < │ %4 = (Base.ult_int)(0x0000000000000000, %3)::Bool
││││╻ & │ %5 = (Base.and_int)(%2, %4)::Bool
.
.
.
│ 11 ┄ %33 = φ (#10 => %28, #34 => %149)::ParticleAmbiguous
│ │ %34 = φ (#10 => %29, #34 => %150)::Int64
│╻ getproperty37 │ %35 = (Base.getfield)(%16, :X)::Any
││ │ %36 = (Base.getfield)(%33, :X)::Any
│ │ %37 = (%35 - %36)::Any
│ 38 │ %38 = (Main.norm)(%37)::Any
.
.
.
Understanding this lowered AST syntax is sort of an art, but you’ll soon learn
that @code_warntype is your best friend when optimizing code. As we scroll
down the AST we see that code encounters types Any in the properties of our
ParticleAmbiguous type, which immediately should raise a red flag to us (Any
is an abstract type). In fact, when running @code_warntype the output
automatically highlights those ::Any asserts in red.
We can take care of those abstract types by defining the properties of the struct parametrically:
In [5]:
"""
This is a particle struct with property types
explicitely/parametrically defined.
"""
struct Particle{T}
# User inputs
X::Array{T, 1} # Position
Gamma::Array{T, 1} # Vectorial circulation
sigma::T # Smoothing radius
# Properties
U::Array{T, 1} # Velocity at particle
J::Array{T, 2} # Jacobian at particle J[i,j]=dUi/dxj
end
# Another initializer alias
Particle{T}(X, Gamma, sigma) where {T} = Particle(X, Gamma, sigma, zeros(T,3), zeros(T, 3, 3))
# Empty initializer
Base.zero(::Type{<:Particle{T}}) where {T} = Particle(zeros(T, 3), zeros(T, 3),
zero(T),
zeros(T, 3), zeros(T, 3, 3))No modifications further are needed in our P2P_pythonic function since Julia’s
multiple dispatch and JIT automatically compiles a version of the function
specialized for our new Particle{T} type on the fly. Still, we will define an
alias to help us compare benchmarks:
In [6]:
P2P_concretetypes(args...) = P2P_pythonic(args...)In [7]:
# Create a 6x6x6 box of concrete Float64 particles
particles = generate_particles(Particle{Float64}, n, lambda)
# Run benchmark
args = (particles, g_wnk, dgdr_wnk)
compare(P2P_concretetypes, P2P_pythonic, args)
@benchmark P2P_concretetypes(args...);P2P_concretetypes is 3.06 times faster than P2P_pythonic (76.488ms vs 233.679ms)
BenchmarkTools.Trial:
memory estimate: 189.93 MiB
allocs estimate: 2275776
--------------
minimum time: 76.488 ms (13.63% GC)
median time: 77.860 ms (13.79% GC)
mean time: 84.567 ms (17.89% GC)
maximum time: 145.918 ms (42.64% GC)
--------------
samples: 12
evals/sample: 1
Voilà! By specifying concrete types in our Particle struct now we have gained
a 3x speed up (we should run @code_warntype again to make sure we got rid of
all abstract types, but I’ll omit it for brevity).
Let’s new see how we compare to C++:
In [8]:
printcomparison(P2P_concretetypes, "C++", false)C++ is 19.14 times faster than P2P_concretetypes (3.996ms vs 76.488ms)
Working with concrete types greatly sped up the computation; however, the C++ version is still ~20x faster than Julia. Let’s see what else can we optimize.
Fix #2: Avoid List Comprehensions
The wonders of list comprehension may tempt you to write some line-efficient code; however, loosely used may lead to a very inefficient computation. Take for example this list-comprehension sum:
In [9]:
sum_list(n) = sum([i for i in 1:n])
@btime sum_list(100); 80.975 ns (1 allocation: 896 bytes)
Here is the version of the same function unrolled without the list comprehension, which does the job 60 times faster:
In [10]:
function sum_unrolled(n)
out = 0
for i in 1:n
out += i
end
return out
end
@btime sum_unrolled(100); 1.374 ns (0 allocations: 0 bytes)
In our P2P function we have a Kronecker delta cross product that we were calculating in just one line as a list comprehension as
# ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp ]
for j in 1:3
Pi.J[:, j] += ( dX[j] / (Pj.sigma*r) * (dgsgmdr*crss) -
gsgm * (3*dX[j]/r^2) * crss -
gsgm * (const4/r^3) * cross([i==j for i in 1:3], Pj.Gamma) )
end
The alternative is to expand it into a few lines as
# ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp ]
for j in 1:3
Pi.J[:, j] += ( dX[j] / (Pj.sigma*r) * (dgsgmdr*crss) -
gsgm * (3*dX[j]/r^2) * crss )
end
# ∂u∂xj(x) = −∑gσ/(4πr^3) δij×Γp
# Adds the Kronecker delta term
aux = -const4 * gsgm / r^3
# j=1
Pi.J[2, 1] -= aux * Pj.Gamma[3]
Pi.J[3, 1] += aux * Pj.Gamma[2]
# j=2
Pi.J[1, 2] += aux * Pj.Gamma[3]
Pi.J[3, 2] -= aux * Pj.Gamma[1]
# j=3
Pi.J[1, 3] -= aux * Pj.Gamma[2]
Pi.J[2, 3] += aux * Pj.Gamma[1]
The problem with list comprehension operations is that it has to allocate memory to build the generated array. Just resist the temptation of using list comprehensions to save yourself a few lines, and simply unroll it. As seen below we get a 1.5x speed up by unrolling this line:
In [11]:
"""
Unrolling the list comprehension
"""
function P2P_nocomprehension(particles, g, dgdr)
for Pi in particles
for Pj in particles
dX = Pi.X - Pj.X
r = norm(dX)
if r != 0
# g_σ and ∂gσ∂r
gsgm = g(r / Pj.sigma)
dgsgmdr = dgdr(r / Pj.sigma)
# K × Γp
crss = cross(-const4 * (dX/r^3), Pj.Gamma)
# U = ∑g_σ(x-xp) * K(x-xp) × Γp
Pi.U[:] += gsgm * crss
# ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp ]
for j in 1:3
Pi.J[:, j] += ( dX[j] / (Pj.sigma*r) * (dgsgmdr*crss) -
gsgm * (3*dX[j]/r^2) * crss )
end
# ∂u∂xj(x) += −∑gσ/(4πr^3) δij×Γp
# Adds the Kronecker delta term
aux = -const4 * gsgm / r^3
# j=1
Pi.J[2, 1] -= aux * Pj.Gamma[3]
Pi.J[3, 1] += aux * Pj.Gamma[2]
# j=2
Pi.J[1, 2] += aux * Pj.Gamma[3]
Pi.J[3, 2] -= aux * Pj.Gamma[1]
# j=3
Pi.J[1, 3] -= aux * Pj.Gamma[2]
Pi.J[2, 3] += aux * Pj.Gamma[1]
end
end
end
endIn [12]:
args = (particles, g_wnk, dgdr_wnk)
compare(P2P_nocomprehension, P2P_concretetypes, args)
@benchmark P2P_nocomprehension(args...);P2P_nocomprehension is 1.52 times faster than P2P_concretetypes (50.196ms vs 76.488ms)
BenchmarkTools.Trial:
memory estimate: 126.16 MiB
allocs estimate: 1300536
--------------
minimum time: 50.196 ms (11.28% GC)
median time: 51.929 ms (11.65% GC)
mean time: 55.319 ms (15.41% GC)
maximum time: 107.039 ms (50.23% GC)
--------------
samples: 19
evals/sample: 1
Fix #3: Reduce Allocation
Next, we notice that the benchmarking test is allotting an unusual amount of
memory (126MiB) and allocation operations (1.3M). I am suspicious that this is
an issue with Julia allowing arrays of dynamic sizes. The first step to solve
this is to do away with creating any internal variables of type arrays. In
the code bellow, notice that I had replaced the array variables dX and crss
with float variables dX1, dX2, dX3, and crss1, crss2, crss3, which leads to
having to fully unroll the inner for-loop:
In [13]:
"""
Reducing memory allocation
"""
function P2P_noallocation(particles, g, dgdr)
for Pi in particles
for Pj in particles
dX1 = Pi.X[1] - Pj.X[1]
dX2 = Pi.X[2] - Pj.X[2]
dX3 = Pi.X[3] - Pj.X[3]
r = norm(Pi.X - Pj.X)
if r != 0
# g_σ and ∂gσ∂r
gsgm = g(r / Pj.sigma)
dgsgmdr = dgdr(r / Pj.sigma)
# K × Γp
crss1, crss2, crss3 = -const4 / r^3 * cross(Pi.X - Pj.X, Pj.Gamma)
# U = ∑g_σ(x-xp) * K(x-xp) × Γp
Pi.U[1] += gsgm * crss1
Pi.U[2] += gsgm * crss2
Pi.U[3] += gsgm * crss3
# ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp ]
# ∂u∂xj(x) += ∑p[(Δxj∂gσ∂r/(σr) − 3Δxjgσ/r^2) K(Δx)×Γp
aux = dgsgmdr/(Pj.sigma*r)* - 3*gsgm /r^2
# j=1
Pi.J[1, 1] += aux * crss1 * dX1
Pi.J[2, 1] += aux * crss2 * dX1
Pi.J[3, 1] += aux * crss3 * dX1
# j=2
Pi.J[1, 2] += aux * crss1 * dX2
Pi.J[2, 2] += aux * crss2 * dX2
Pi.J[3, 2] += aux * crss3 * dX2
# j=3
Pi.J[1, 3] += aux * crss1 * dX3
Pi.J[2, 3] += aux * crss2 * dX3
Pi.J[3, 3] += aux * crss3 * dX3
# Adds the Kronecker delta term
aux = -const4 * gsgm / r^3
# j=1
Pi.J[2, 1] -= aux * Pj.Gamma[3]
Pi.J[3, 1] += aux * Pj.Gamma[2]
# j=2
Pi.J[1, 2] += aux * Pj.Gamma[3]
Pi.J[3, 2] -= aux * Pj.Gamma[1]
# j=3
Pi.J[1, 3] -= aux * Pj.Gamma[2]
Pi.J[2, 3] += aux * Pj.Gamma[1]
end
end
end
endIn [14]:
args = (particles, g_wnk, dgdr_wnk)
compare(P2P_noallocation, P2P_nocomprehension, args)
@benchmark P2P_noallocation(args...);P2P_noallocation is 3.68 times faster than P2P_nocomprehension (13.627ms vs 50.196ms)
BenchmarkTools.Trial:
memory estimate: 21.99 MiB
allocs estimate: 325296
--------------
minimum time: 13.627 ms (7.41% GC)
median time: 15.615 ms (8.40% GC)
mean time: 16.582 ms (12.83% GC)
maximum time: 59.011 ms (66.91% GC)
--------------
samples: 61
evals/sample: 1
Here we have reduced the memory allocated from 126MiB to 22MiB, leading to a 3.5x speed up. Let’s see what else can we do to decrease memory allocation.
Fix #4: No Linear Algebra
The next thing to consider is that doing linear algebra operations
using the Julia base library (i.e., dot(X,X),
cross(X,X), norm(X,X)) is more expensive that explicitely unfolding the
operation into lines of code. I am suspicious that this is a memory allocation
problem since these functions need to allocate internal arrays to store the
computation prior to outputting the result. Here is the code without any
functional linear algebra functions from the base library (notice that I no longer use norm() nor
cross()):
In [15]:
"""
No linear algebra functions
"""
function P2P_nolinalg(particles, g, dgdr)
for Pi in particles
for Pj in particles
dX1 = Pi.X[1] - Pj.X[1]
dX2 = Pi.X[2] - Pj.X[2]
dX3 = Pi.X[3] - Pj.X[3]
r = sqrt(dX1*dX1 + dX2*dX2 + dX3*dX3)
if r != 0
# g_σ and ∂gσ∂r
gsgm = g(r / Pj.sigma)
dgsgmdr = dgdr(r / Pj.sigma)
# K × Γp
crss1 = -const4 / r^3 * ( dX2*Pj.Gamma[3] - dX3*Pj.Gamma[2] )
crss2 = -const4 / r^3 * ( dX3*Pj.Gamma[1] - dX1*Pj.Gamma[3] )
crss3 = -const4 / r^3 * ( dX1*Pj.Gamma[2] - dX2*Pj.Gamma[1] )
# U = ∑g_σ(x-xp) * K(x-xp) × Γp
Pi.U[1] += gsgm * crss1
Pi.U[2] += gsgm * crss2
Pi.U[3] += gsgm * crss3
# ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp ]
# ∂u∂xj(x) += ∑p[(Δxj∂gσ∂r/(σr) − 3Δxjgσ/r^2) K(Δx)×Γp
aux = dgsgmdr/(Pj.sigma*r)* - 3*gsgm /r^2
# j=1
Pi.J[1, 1] += aux * crss1 * dX1
Pi.J[2, 1] += aux * crss2 * dX1
Pi.J[3, 1] += aux * crss3 * dX1
# j=2
Pi.J[1, 2] += aux * crss1 * dX2
Pi.J[2, 2] += aux * crss2 * dX2
Pi.J[3, 2] += aux * crss3 * dX2
# j=3
Pi.J[1, 3] += aux * crss1 * dX3
Pi.J[2, 3] += aux * crss2 * dX3
Pi.J[3, 3] += aux * crss3 * dX3
# Adds the Kronecker delta term
aux = -const4 * gsgm / r^3
# j=1
Pi.J[2, 1] -= aux * Pj.Gamma[3]
Pi.J[3, 1] += aux * Pj.Gamma[2]
# j=2
Pi.J[1, 2] += aux * Pj.Gamma[3]
Pi.J[3, 2] -= aux * Pj.Gamma[1]
# j=3
Pi.J[1, 3] -= aux * Pj.Gamma[2]
Pi.J[2, 3] += aux * Pj.Gamma[1]
end
end
end
endIn [16]:
args = (particles, g_wnk, dgdr_wnk)
compare(P2P_nolinalg, P2P_noallocation, args)
@benchmark P2P_nolinalg(args...);P2P_nolinalg is 2.10 times faster than P2P_noallocation (6.487ms vs 13.627ms)
BenchmarkTools.Trial:
memory estimate: 0 bytes
allocs estimate: 0
--------------
minimum time: 6.487 ms (0.00% GC)
median time: 7.140 ms (0.00% GC)
mean time: 7.198 ms (0.00% GC)
maximum time: 9.172 ms (0.00% GC)
--------------
samples: 139
evals/sample: 1
By doing away with linear algebra functions we are now not allocating any memory, reaching an extra 2x speed up.
Fix #5: Fine Tuning
Notice that by now we are achieving benchmarks in the same order of magnitude than C++ (6.5ms in Julia vs 4.0ms in C++):
In [17]:
printcomparison(P2P_nolinalg, "C++", false)C++ is 1.62 times faster than P2P_nolinalg (3.996ms vs 6.487ms)
What we did prior to this point were general principles that apply to any code that attempts to get high performance. What it follows now is fine tuning of our code in ways that only apply to the specific computation that we are performing.
For instance, recall that our P2P function receives any user-defined regularizing kernel that our function calls through this lines:
function P2P(sources, targets, g::Function, dgdr::Function)
for Pi in targets
for Pj in sources
.
.
.
# g_σ and ∂gσ∂r
gsgm = g(r / Pj.sigma)
dgsgmdr = dgdr(r / Pj.sigma)
.
.
.
end
end
end
For the case of Winckelmans’ kernel, g and dgdr look like this:
g_wnk(r) = r^3 * (r^2 + 2.5) / (r^2 + 1)^2.5
dgdr_wnk(r) = 7.5 * r^2 / (r^2 + 1)^3.5
We notice that each of these function calculate a power operation independently,
(r^2 + 1)^2.5 and (r^2 + 1)^3.5. I have observed that any sort of non-integer
power operation takes Julia more than any basic arithmetic operation or
even space allocation. Thus, we can save computation by merging this two functions
and reusing the square root calculation as
In [45]:
function g_dgdr_wnk(r)
aux = (r^2 + 1)^2.5
# Returns g and dgdr
return r^3*(r^2 + 2.5)/aux, 7.5*r^2/(aux*(r^2 + 1))
end
"""
Reuses sqrt, reducing to only one power calculation
"""
function P2P_reusesqrt(particles, g_dgdr)
for Pi in particles
for Pj in particles
dX1 = Pi.X[1] - Pj.X[1]
dX2 = Pi.X[2] - Pj.X[2]
dX3 = Pi.X[3] - Pj.X[3]
r = sqrt(dX1*dX1 + dX2*dX2 + dX3*dX3)
if r != 0
# g_σ and ∂gσ∂r
gsgm, dgsgmdr = g_dgdr(r / Pj.sigma)
# K × Γp
crss1 = -const4 / r^3 * ( dX2*Pj.Gamma[3] - dX3*Pj.Gamma[2] )
crss2 = -const4 / r^3 * ( dX3*Pj.Gamma[1] - dX1*Pj.Gamma[3] )
crss3 = -const4 / r^3 * ( dX1*Pj.Gamma[2] - dX2*Pj.Gamma[1] )
# U = ∑g_σ(x-xp) * K(x-xp) × Γp
Pi.U[1] += gsgm * crss1
Pi.U[2] += gsgm * crss2
Pi.U[3] += gsgm * crss3
# ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp ]
# ∂u∂xj(x) += ∑p[(Δxj∂gσ∂r/(σr) − 3Δxjgσ/r^2) K(Δx)×Γp
aux = dgsgmdr/(Pj.sigma*r)* - 3*gsgm /r^2
# j=1
Pi.J[1, 1] += aux * crss1 * dX1
Pi.J[2, 1] += aux * crss2 * dX1
Pi.J[3, 1] += aux * crss3 * dX1
# j=2
Pi.J[1, 2] += aux * crss1 * dX2
Pi.J[2, 2] += aux * crss2 * dX2
Pi.J[3, 2] += aux * crss3 * dX2
# j=3
Pi.J[1, 3] += aux * crss1 * dX3
Pi.J[2, 3] += aux * crss2 * dX3
Pi.J[3, 3] += aux * crss3 * dX3
# Adds the Kronecker delta term
aux = -const4 * gsgm / r^3
# j=1
Pi.J[2, 1] -= aux * Pj.Gamma[3]
Pi.J[3, 1] += aux * Pj.Gamma[2]
# j=2
Pi.J[1, 2] += aux * Pj.Gamma[3]
Pi.J[3, 2] -= aux * Pj.Gamma[1]
# j=3
Pi.J[1, 3] -= aux * Pj.Gamma[2]
Pi.J[2, 3] += aux * Pj.Gamma[1]
end
end
end
endIn [19]:
args = (particles, g_dgdr_wnk)
compare(P2P_reusesqrt, P2P_nolinalg, args)
@benchmark P2P_reusesqrt(args...);P2P_reusesqrt is 1.60 times faster than P2P_nolinalg (4.050ms vs 6.487ms)
BenchmarkTools.Trial:
memory estimate: 0 bytes
allocs estimate: 0
--------------
minimum time: 4.050 ms (0.00% GC)
median time: 4.479 ms (0.00% GC)
mean time: 4.493 ms (0.00% GC)
maximum time: 5.361 ms (0.00% GC)
--------------
samples: 223
evals/sample: 1
Hence, by simply avoiding one extra power calculation we have now gained a 1.6x speed up.
Final Fix: @inbounds, @simd, @fastmath
This is straight from the Julia official documentation:
Like many modern programming languages, Julia uses bounds checking to ensure program safety when accessing arrays. In tight inner loops or other performance critical situations, you may wish to skip these bounds checks to improve runtime performance. For instance, in order to emit vectorized (SIMD) instructions, your loop body cannot contain branches, and thus cannot contain bounds checks. Consequently, Julia includes an @inbounds(…) macro to tell the compiler to skip such bounds checks within the given block. User-defined array types can use the @boundscheck(…) macro to achieve context-sensitive code selection.
Write @simd in front of for loops to promise that the iterations are independent and may be reordered. Note that in many cases, Julia can automatically vectorize code without the @simd macro; it is only beneficial in cases where such a transformation would otherwise be illegal, including cases like allowing floating-point re-associativity and ignoring dependent memory accesses (@simd ivdep). Again, be very careful when asserting @simd as erroneously annotating a loop with dependent iterations may result in unexpected results. In particular, note that setindex! on some AbstractArray subtypes is inherently dependent upon iteration order. This feature is experimental and could change or disappear in future versions of Julia.
Use @fastmath to allow floating point optimizations that are correct for real numbers, but lead to differences for IEEE numbers. Be careful when doing this, as this may change numerical results. This corresponds to the -ffast-math option of clang.
I won’t dive into details, but what I have realized is that you can get a speed
up from the three macros listed above only when you are working with data
structures that pass the isbits
test. In practice,
that means that our structs can’t contain any arrays, which lead us to
redefining the struct as follows:
In [30]:
struct ParticleNoArr{T}
# User inputs
X1::T
X2::T
X3::T
Gamma1::T
Gamma2::T
Gamma3::T
sigma::T
end
# Empty initializer
Base.zero(::Type{<:ParticleNoArr{T}}) where {T} = ParticleNoArr(zeros(T, 7)...)
# Another initializer alias
ParticleNoArr{T}(X, Gamma, sigma) where T = ParticleNoArr(X..., Gamma..., sigma);With that twist, now we implement @simd in the internal for loop, while both
@fastmath and @inbounds wrap all floating point operations and output
allocation:
In [50]:
"""
Implementation of @simd, @fastmath, and @inbounds
"""
function P2P_FINAL(particles, g_dgdr, U, J)
for (i, Pi) in enumerate(particles)
@simd for Pj in particles
@fastmath @inbounds begin
dX1 = Pi.X1 - Pj.X1
dX2 = Pi.X2 - Pj.X2
dX3 = Pi.X3 - Pj.X3
r = sqrt(dX1*dX1 + dX2*dX2 + dX3*dX3)
if r != 0
# g_σ and ∂gσ∂r
gsgm, dgsgmdr = g_dgdr(r / Pj.sigma)
# K × Γp
crss1 = -const4 / r^3 * ( dX2*Pj.Gamma3 - dX3*Pj.Gamma2 )
crss2 = -const4 / r^3 * ( dX3*Pj.Gamma1 - dX1*Pj.Gamma3 )
crss3 = -const4 / r^3 * ( dX1*Pj.Gamma2 - dX2*Pj.Gamma1 )
# U = ∑g_σ(x-xp) * K(x-xp) × Γp
U[1, i] += gsgm * crss1
U[2, i] += gsgm * crss2
U[3, i] += gsgm * crss3
# ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp ]
# ∂u∂xj(x) += ∑p[(Δxj∂gσ∂r/(σr) − 3Δxjgσ/r^2) K(Δx)×Γp
aux = dgsgmdr/(Pj.sigma*r)* - 3*gsgm /r^2
# j=1
J[1, 1, i] += aux * crss1 * dX1
J[2, 1, i] += aux * crss2 * dX1
J[3, 1, i] += aux * crss3 * dX1
# j=2
J[1, 2, i] += aux * crss1 * dX2
J[2, 2, i] += aux * crss2 * dX2
J[3, 2, i] += aux * crss3 * dX2
# j=3
J[1, 3, i] += aux * crss1 * dX3
J[2, 3, i] += aux * crss2 * dX3
J[3, 3, i] += aux * crss3 * dX3
# Adds the Kronecker delta term
aux = -const4 * gsgm / r^3
# j=1
J[2, 1, i] -= aux * Pj.Gamma3
J[3, 1, i] += aux * Pj.Gamma2
# j=2
J[1, 2, i] += aux * Pj.Gamma3
J[3, 2, i] -= aux * Pj.Gamma1
# j=3
J[1, 3, i] -= aux * Pj.Gamma2
J[2, 3, i] += aux * Pj.Gamma1
end
end
end
end
endIn [51]:
# Create a 6x6x6 box of particles of our new ParticleNoArr struct
particlesNoArr = generate_particles(ParticleNoArr{Float64}, n, lambda)
# Pre-allocate outputs in separate arrays
U = zeros(Float64, 3, length(particlesNoArr) )
J = zeros(Float64, 3, 3, length(particlesNoArr) )
# Run benchmark
args = (particlesNoArr, g_dgdr_wnk, U, J)
compare(P2P_FINAL, P2P_reusesqrt, args)
@benchmark P2P_FINAL(args...);P2P_FINAL is 1.14 times faster than P2P_reusesqrt (3.552ms vs 4.050ms)
BenchmarkTools.Trial:
memory estimate: 0 bytes
allocs estimate: 0
--------------
minimum time: 3.552 ms (0.00% GC)
median time: 4.090 ms (0.00% GC)
mean time: 4.056 ms (0.00% GC)
maximum time: 4.761 ms (0.00% GC)
--------------
samples: 247
evals/sample: 1
Comparing to C++, our Julia code is now as fast —and a little faster—than C++:
In [52]:
printcomparison(P2P_FINAL, "C++", true)P2P_FINAL is 1.12 times faster than C++ (3.552ms vs 3.996ms)
However, our Julia code is using clang’s fastmath compilation mode. A more fair
comparison is done by also compiling C++ with the -ffast-math flag in addition
to -O3, which I have coded, compiled, and wrapped in this
CxxWrap:
In [34]:
# Run C++ benchmark with -ffast-math flag
cpptime = CxxVortexTestFASTMATH.benchmarkP2P_wrap(ntests, n, Float32(lambda), verbose)
# Store benchmark result
benchtime["C++ -ffast-math"] = cpptime;Samples: 1000
min time: 1.40114 ms
ave time: 1.72617 ms
max time: 2.71737 ms
In [53]:
printcomparison(P2P_FINAL, "C++ -ffast-math", false)C++ -ffast-math is 2.54 times faster than P2P_FINAL (1.401ms vs 3.552ms)
And once again, -ffast-math places C++ at a modest 2.5x over the optimized
Julia code; but be not discouraged, we are talking about a dynamic-like language
that is only 0.4x slower than C++! =]
NOTE: In a subsequent test I commented out the two power operations (r =
sqrt(...) and gsgm, dgsgmdr = g_dgdr(r/Pj.sigma)) and the resulting wall-
clock time went down from 6.35ms to 0.496ms, meaning that at this stage the
overhead is on non-algebraic operations and that the function could further be
optimized only by finding a more efficient way of calculation powers in Julia.
Julia as Fast as C++
Finally, let’s take a second to compare where we started and where we ended. Our initial Julia function was very compact and understandable, however it was more than 50x slower than its C++ counterpart:
"""
Pythonic programming approach
"""
function P2P_pythonic(particles, g, dgdr)
for Pi in particles
for Pj in particles
dX = Pi.X - Pj.X
r = norm(dX)
if r != 0
# g_σ and ∂gσ∂r
gsgm = g(r / Pj.sigma)
dgsgmdr = dgdr(r / Pj.sigma)
# K × Γp
crss = cross(-const4 * (dX/r^3), Pj.Gamma)
# U = ∑g_σ(x-xp) * K(x-xp) × Γp
Pi.U[:] += gsgm * crss
# ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp ]
for j in 1:3
Pi.J[:, j] += ( dX[j] / (Pj.sigma*r) * (dgsgmdr*crss) -
gsgm * (3*dX[j]/r^2) * crss -
gsgm * (const4/r^3) * cross([i==j for i in 1:3], Pj.Gamma) )
end
end
end
end
end
In [48]:
printcomparison(P2P_pythonic, "C++", false)C++ is 58.48 times faster than P2P_pythonic (3.996ms vs 233.679ms)
After all the optimization we end up with about twice the amount of lines, but
65x faster that the original version and only 2.5x slower than C++ with
-ffastmath:
"""
Optimized Julia code
"""
function P2P_FINAL(particles, g_dgdr, U, J)
for (i, Pi) in enumerate(particles)
@simd for Pj in particles
@fastmath @inbounds begin
dX1 = Pi.X1 - Pj.X1
dX2 = Pi.X2 - Pj.X2
dX3 = Pi.X3 - Pj.X3
r = sqrt(dX1*dX1 + dX2*dX2 + dX3*dX3)
if r != 0
# g_σ and ∂gσ∂r
gsgm, dgsgmdr = g_dgdr(r / Pj.sigma)
# K × Γp
crss1 = -const4 / r^3 * ( dX2*Pj.Gamma3 - dX3*Pj.Gamma2 )
crss2 = -const4 / r^3 * ( dX3*Pj.Gamma1 - dX1*Pj.Gamma3 )
crss3 = -const4 / r^3 * ( dX1*Pj.Gamma2 - dX2*Pj.Gamma1 )
# U = ∑g_σ(x-xp) * K(x-xp) × Γp
U[1, i] += gsgm * crss1
U[2, i] += gsgm * crss2
U[3, i] += gsgm * crss3
# ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp ]
# ∂u∂xj(x) += ∑p[(Δxj∂gσ∂r/(σr) − 3Δxjgσ/r^2) K(Δx)×Γp
aux = dgsgmdr/(Pj.sigma*r)* - 3*gsgm /r^2
# j=1
J[1, 1, i] += aux * crss1 * dX1
J[2, 1, i] += aux * crss2 * dX1
J[3, 1, i] += aux * crss3 * dX1
# j=2
J[1, 2, i] += aux * crss1 * dX2
J[2, 2, i] += aux * crss2 * dX2
J[3, 2, i] += aux * crss3 * dX2
# j=3
J[1, 3, i] += aux * crss1 * dX3
J[2, 3, i] += aux * crss2 * dX3
J[3, 3, i] += aux * crss3 * dX3
# Adds the Kronecker delta term
aux = -const4 * gsgm / r^3
# j=1
J[2, 1, i] -= aux * Pj.Gamma3
J[3, 1, i] += aux * Pj.Gamma2
# j=2
J[1, 2, i] += aux * Pj.Gamma3
J[3, 2, i] -= aux * Pj.Gamma1
# j=3
J[1, 3, i] -= aux * Pj.Gamma2
J[2, 3, i] += aux * Pj.Gamma1
end
end
end
end
end
In [54]:
printcomparison(P2P_FINAL, P2P_pythonic, true)
printcomparison(P2P_FINAL, "C++", true)
printcomparison(P2P_FINAL, "C++ -ffast-math", false)P2P_FINAL is 65.79 times faster than P2P_pythonic (3.552ms vs 233.679ms)
P2P_FINAL is 1.12 times faster than C++ (3.552ms vs 3.996ms)
C++ -ffast-math is 2.54 times faster than P2P_FINAL (1.401ms vs 3.552ms)
Oddly enough, through the optimization process we naturally morphed the code into something that looks very similar to the C++ version:
// Particle-to-particle interactions
void P2P(Particle * P, const int numParticles) {
real_t r, ros, aux, g_sgm, dgdr;
vec3 dX;
for (int i=0; i<numParticles; i++) {
for (int j=0; j<numParticles; j++) {
dX[0] = P[i].X[0] - P[j].X[0];
dX[1] = P[i].X[1] - P[j].X[1];
dX[2] = P[i].X[2] - P[j].X[2];
r = sqrt(dX[0]*dX[0] + dX[1]*dX[1] + dX[2]*dX[2]);
if (r!=0){
ros = r/P[j].sigma;
// Evaluate g_σ and ∂gσ∂r
aux = pow(ros*ros + 1.0, 2.5);
g_sgm = ros*ros*ros * (ros*ros + 2.5) / aux;
dgdr = 7.5 * ros*ros / ( aux * (ros*ros + 1.0) );
// u(x) = ∑g_σ(x−xp) K(x−xp) × Γp
aux = (- const4 / (r*r*r)) * g_sgm;
P[i].U[0] += ( dX[1]*P[j].Gamma[2] - dX[2]*P[j].Gamma[1] ) * aux;
P[i].U[1] += ( dX[2]*P[j].Gamma[0] - dX[0]*P[j].Gamma[2] ) * aux;
P[i].U[2] += ( dX[0]*P[j].Gamma[1] - dX[1]*P[j].Gamma[0] ) * aux;
// ∂u∂xj(x) = ∑[ ∂gσ∂xj(x−xp) * K(x−xp)×Γp + gσ(x−xp) * ∂K∂xj(x−xp)×Γp]
aux = (- const4 / (r*r*r)) * (dgdr/(P[j].sigma*r) - 3.0*g_sgm/(r*r));
for (int k=0; k<3; k++){
P[i].J[3*k + 0] += ( dX[1]*P[j].Gamma[2] - dX[2]*P[j].Gamma[1] )*aux*dX[k];
P[i].J[3*k + 1] += ( dX[2]*P[j].Gamma[0] - dX[0]*P[j].Gamma[2] )*aux*dX[k];
P[i].J[3*k + 2] += ( dX[0]*P[j].Gamma[1] - dX[1]*P[j].Gamma[0] )*aux*dX[k];
}
// Adds the Kronecker delta term
aux = - const4 * g_sgm / (r*r*r);
// k=1
P[i].J[3*0 + 1] -= aux * P[j].Gamma[2];
P[i].J[3*0 + 2] += aux * P[j].Gamma[1];
// k=2
P[i].J[3*1 + 0] += aux * P[j].Gamma[2];
P[i].J[3*1 + 2] -= aux * P[j].Gamma[0];
// k=3
P[i].J[3*2 + 0] -= aux * P[j].Gamma[1];
P[i].J[3*2 + 1] += aux * P[j].Gamma[0];
}
}
}
}
In conclusion, if you are trying to get a piece of code fit for high-performance computing, my recommendation is to forget about elegant and line-efficient coding (“pythonic” some would say), and code in C++-esque style from the beginning.
Conclusions
-
Without foreknowledge of the types to be handled, Julia can’t optimize the code during compilation. Multiple dispatch and JIT will compile a well-defined version of every function based on the arguments that is given on the fly, but this is not the case for structs properties. Always define your structs with its properties explicitely specified as concrete types.
-
@code_warntypeis your best friend when trying to catch abstract types in your code. -
Avoid memory allocation: If your function is allocating an insane amount of memory, that’s an indication that you are saving yourself some lines of code while sacrificing computation efficiency (e.g., inline list-comprehension operations or array algebra instead of unrolling the operations into explicit code). I recommend using
@timeor@benchmarkto check memory allocation. -
Avoid storing intermediate calculations in internal arrays, just use Float variables. For some reason Julia spends a lot of time trying to allocate space for internal arrays.
-
Any non-integer power operations (
^orsqrt) are very expensive in Julia. If possible, reduce these operations by storing and reusing previous calculations. -
At all cost, avoid using functions from the LinearAlgebra package (like
dot,cross,norm) as they will require allocating memory for internal arrays. -
For some reason,
@simd,@fastmath, and@inboundsspeed up your code only when working withisbitstypes. -
Forget about elegant and line-efficient coding (“pythonic” some would say), and code in C++-esque style in parts of the code where performance is critical.
Notes
- This benchmark was done in a Dell Latitude 5580 laptop (Intel® Core™ i7-7820HQ CPU @ 2.90GHz × 8 ) in only one process.
- Julia v1.0.3 was used.
- The official Julia documentation also provide very useful tips for performance.
- The code used in this post is available here: https://github.com/EdoAlvarezR/post-juliavscpp
References
- Winckelmans, G. S., & Leonard, A. (1993). Contributions to Vortex Particle Methods for the Computation of Three-Dimensional Incompressible Unsteady Flows. Journal of Computational Physics, 109(2), 247–273. https://doi.org/10.1006/jcph.1993.1216
- Alvarez, E. J., & Ning, A. (2018). Development of a Vortex Particle Code for the Modeling of Wake Interaction in Distributed Propulsion. In 2018 Applied Aerodynamics Conference (pp. 1–22). American Institute of Aeronautics and Astronautics. https://doi.org/10.2514/6.2018-3646 . PDF Available
