Fluid Domain
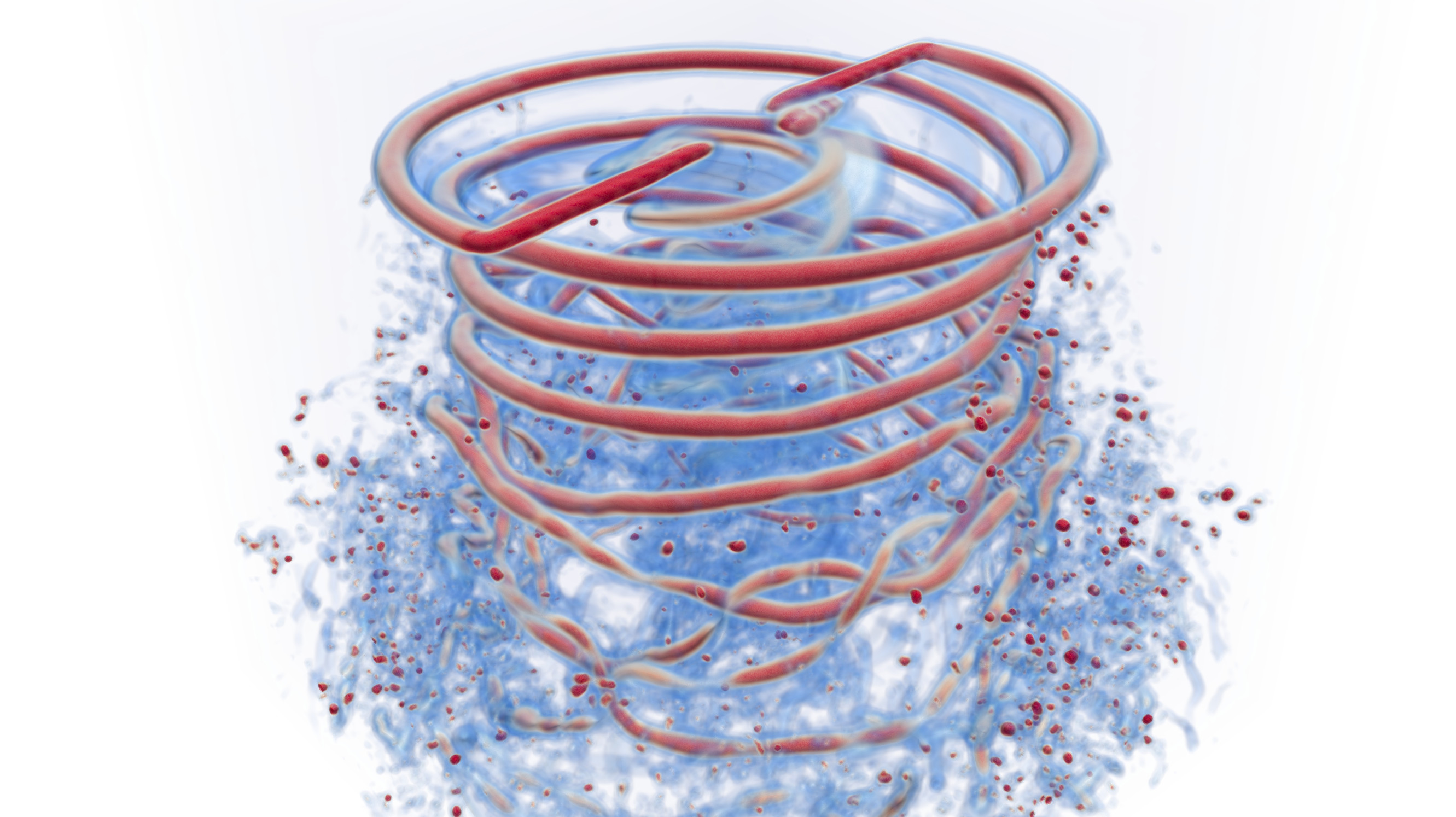
The full fluid domain can be computed in a postprocessing step from the particle field. This is possible because the particle field is a radial basis function that constructs the vorticity field, and the velocity field can be recovered from vorticity through the Biot-Savart law.
Here we show how to use uns.computefluiddomain to read a simulation and process it to generate its fluid domain.
#=##############################################################################
Computes the fluid domain of DJI 9443 simulation using a volumetric domain.
This is done probing the velocity and vorticity that the particle field
induces at each node of a Cartesian grids.
NOTE: The fluid domain generated here does not include the freestream
velocity, which needs to be added manually inside ParaView (if any).
=###############################################################################
import FLOWUnsteady as uns
import FLOWUnsteady: vpm, gt, dot, norm
# --------------- INPUTS AND OUTPUTS -------------------------------------------
# INPUT OPTIONS
simulation_name = "rotorhover-example-midhigh00" # Simulation to read
read_path = "/home/edoalvar/simulationdata202330/"*simulation_name # Where to read simulation from
pfield_prefix = "singlerotor_pfield" # Prefix of particle field files to read
staticpfield_prefix = "singlerotor_staticpfield" # Prefix of static particle field files to read
nums = [719] # Time steps to process
# OUTPUT OPTIONS
save_path = joinpath(read_path, "..", simulation_name*"-fdom") # Where to save fluid domain
output_prefix = "singlerotor" # Prefix of output files
prompt = true # Whether to prompt the user
verbose = true # Enable verbose
v_lvl = 0 # Verbose indentation level
# -------------- PARAMETERS ----------------------------------------------------
# Simulation information
R = 0.12 # (m) rotor radius
AOA = 0.0 # (deg) angle of attack or incidence angle
# Grid
L = R # (m) reference length
dx, dy, dz = L/50, L/50, L/50 # (m) cell size in each direction
Pmin = L*[-0.50, -1.25, -1.25] # (m) minimum bounds
Pmax = L*[ 2.00, 1.25, 1.25] # (m) maximum bounds
NDIVS = ceil.(Int, (Pmax .- Pmin)./[dx, dy, dz]) # Number of cells in each direction
nnodes = prod(NDIVS .+ 1) # Total number of nodes
Oaxis = gt.rotation_matrix2(0, 0, AOA) # Orientation of grid
# VPM settings
maxparticles = Int(1.0e6 + nnodes) # Maximum number of particles
fmm = vpm.FMM(; p=4, ncrit=50, theta=0.4, nonzero_sigma=false, ε_tol=nothing) # FMM parameters
scale_sigma = 1.00 # Shrink smoothing radii by this factor
f_sigma = 0.5 # Smoothing of node particles as sigma = f_sigma*meansigma
maxsigma = L/10 # Particles larger than this get shrunk to this size (this helps speed up computation)
maxmagGamma = Inf # Any vortex strengths larger than this get clipped to this value
include_staticparticles = true # Whether to include the static particles embedded in the solid surfaces
other_file_prefs = include_staticparticles ? [staticpfield_prefix] : []
other_read_paths = [read_path for i in 1:length(other_file_prefs)]
if verbose
println("\t"^(v_lvl)*"Fluid domain grid")
println("\t"^(v_lvl)*"NDIVS =\t$(NDIVS)")
println("\t"^(v_lvl)*"Number of nodes =\t$(nnodes)")
end
# --------------- PROCESSING SETUP ---------------------------------------------
if verbose
println("\t"^(v_lvl)*"Getting ready to process $(read_path)")
println("\t"^(v_lvl)*"Results will be saved under $(save_path)")
end
# Create save path
if save_path != read_path
gt.create_path(save_path, prompt)
end
# Generate function to process the field clipping particle sizes
preprocessing_pfield = uns.generate_preprocessing_fluiddomain_pfield(maxsigma, maxmagGamma;
verbose=verbose, v_lvl=v_lvl+1)
# --------------- PROCESS SIMULATION -------------------------------------------
nthreads = 1 # Total number of threads
nthread = 1 # Number of this thread
dnum = floor(Int, length(nums)/nthreads) # Number of time steps per thread
threaded_nums = [view(nums, dnum*i+1:(i<nthreads-1 ? dnum*(i+1) : length(nums))) for i in 0:nthreads-1]
for these_nums in threaded_nums[nthread:nthread]
uns.computefluiddomain( Pmin, Pmax, NDIVS,
maxparticles,
these_nums, read_path, pfield_prefix;
Oaxis=Oaxis,
fmm=fmm,
f_sigma=f_sigma,
save_path=save_path,
file_pref=output_prefix, grid_names=["_fdom"],
other_file_prefs=other_file_prefs,
other_read_paths=other_read_paths,
userfunction_pfield=preprocessing_pfield,
verbose=verbose, v_lvl=v_lvl)
end

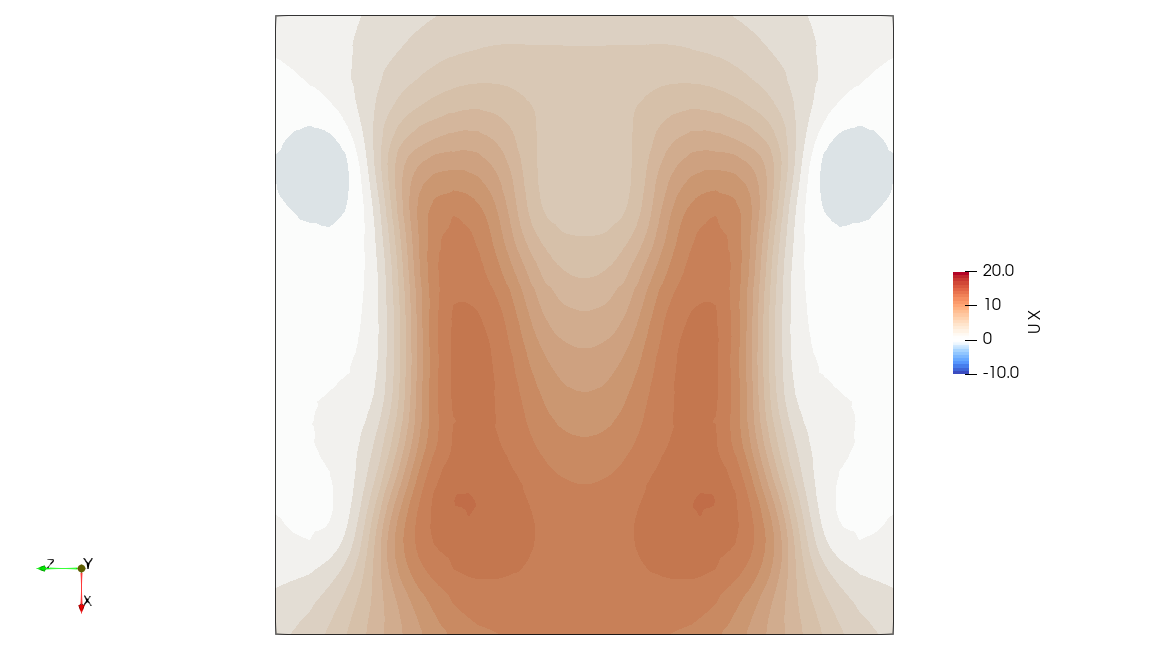
High Fidelity
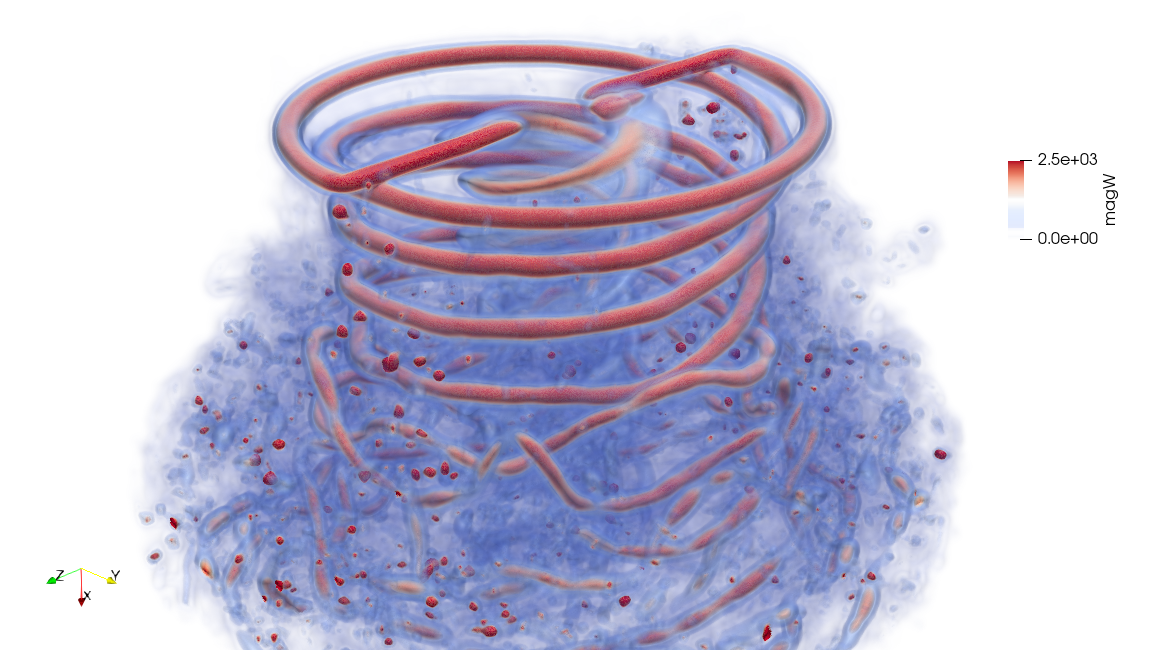
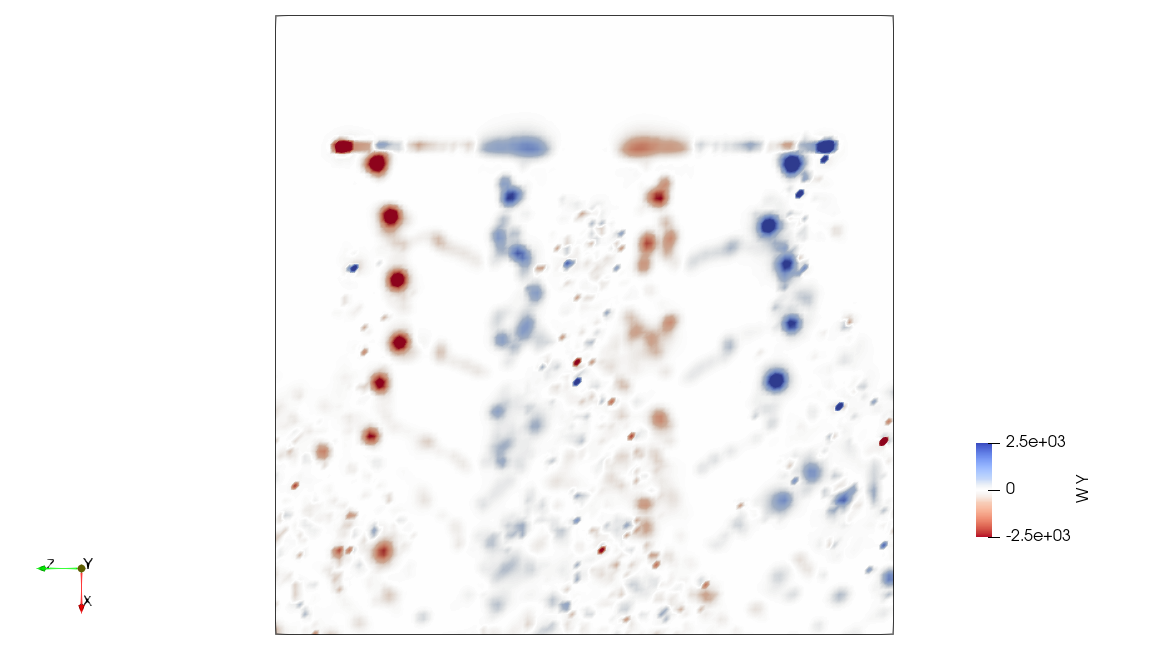
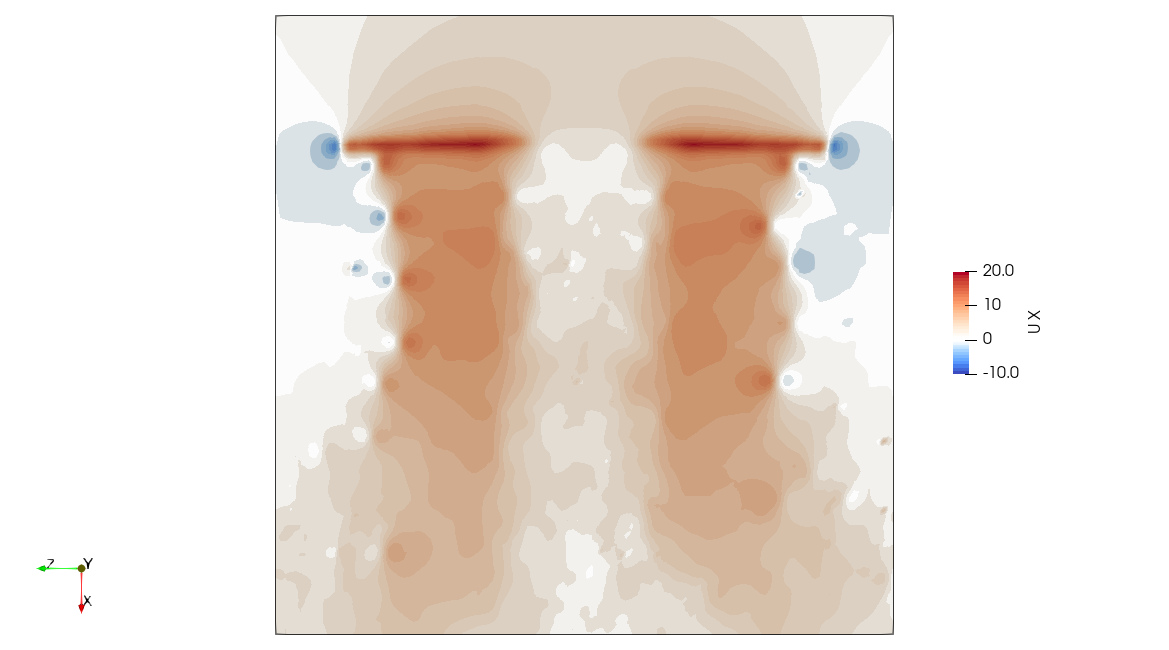
The .pvsm files visualizing the fluid domain as shown above are available in the following links
(right click → save as...).
To open in ParaView: File → Load State → (select .pvsm file) then select "Search files under specified directory" and point it to the folder where the simulation was saved.